The Métabolome et Valorisation de la Biodiversité Végétale (MVBV) team of the Institut de Chimie de Nice (Université Côte d’Azur, France) is directed by Prof. Xavier Fernandez who works in the field of natural products analysis. Together with BotaniCert, an analytical laboratory directed by Dr. Francis Hadji-Minaglou and specialized in the analysis and authentication of plant material, the MVBV team has been working on a collaborative project to find new natural preservatives in Mediterranean plant species for the cosmetic industry.
Introduction
Plant extracts offer a large source of bioactive molecules. Most often, the screening for bioactivity is performed with the dilution method in 96 well plates. The major disadvantage of this method is the lack of information on the chemical class and number of active constituents of the extract. HPTLC is a widely employed technique for testing of natural products, e.g. identification and test for adulteration. Since the last decade, a growing interest in HPTLC hyphenated with bioassays (HPTLC-EDA) has been noted [1].
For a simplified identification of active ingredients in plant extracts we have chosen HPTLC bioautography. Due to the combination of chromatographic separation and effect-directed analyses, HPTLC allows to directly localize active ingredients, avoiding a time-consuming bio-guided fractionation. Up to 15 extracts can be analyzed at the same time. In our study, Aspergillus brasiliensis has been selected to screen for antifungal active ingredients. Positive compounds can be promising candidates as natural preservatives for cosmetic products.
Standard solutions
Glycyrrhizic acid, quercetin and rutin (0.5 mg/mL in methanol)
Sample preparation
Dry plant material is grounded and macerated in ethanol 75% (plant volume/solvent volume 1:10) for 2 h at room temperature. After centrifugation the supernatants are used as test solutions.
Chromatogram layer
HPTLC plate silica gel 60 F254 (Merck), 20 x 10 cm
Sample application
Automatic TLC Sampler (ATS 4), application as bands, 15 tracks, band length 8.0 mm, distance from left edge 20.0 mm, distance from lower edge 8.0 mm, application volume 20.0 μL for samples and 2.0 μL for standards
Chromatography
In the ADC 2 with chamber saturation with filter paper 20 min and after activation at 33% relative humidity for 10 min using a saturated solution of magnesium chloride, development with ethyl acetate – water – acetic acid – formic acid 100: 26:11:11 to the migration distance of 70 mm (from the lower edge), drying for 5 min
Post-chromatographic derivatization
- The plate is immersed into anisaldehyde-sulfuric acid reagent (10 mL sulfuric acid is added to 170 mL methanol, 20 mL acetic acid and then 1 mL anisaldehyde is added to that solution) using the Chromatogram Immersion Device (immersion speed 6cm/s, immersion time 0s), dried for 30s with cold air, and heating at 100 °C for 5 min using the TLC Plate Heater.
- The plate is heated for 3min at 100°C using the TLC Plate Heater then immersed in Natural product (NP) reagent (1 g diphenylborinic acid aminoethylester in 200 mL ethyl acetate) and in PEG solution (10 g polyethylene glycol 400 in 200mL dichloromethane) using the Chromatogram Immersion Device (immersion speed 6cm/s, immersion time 0 s).
Editor’s Note: in many cases a sequential derivatization with first NP reagent and then with anisaldehyde reagent on the same plate is possible. The fluorescence enhancer (PEG) is then not used.
Bioautography with Aspergillus brasiliensis
The plate is conditioned with the culture broth (Sabouraud), then manually sprayed with a suspension of spores of A. brasiliensis at 105 spores/mL, and incubated at room temperature (25°C) 48 h to 72 h.
Documentation
TLC Visualizer under UV 254 nm, UV 366 nm, and white light
Results and discussion
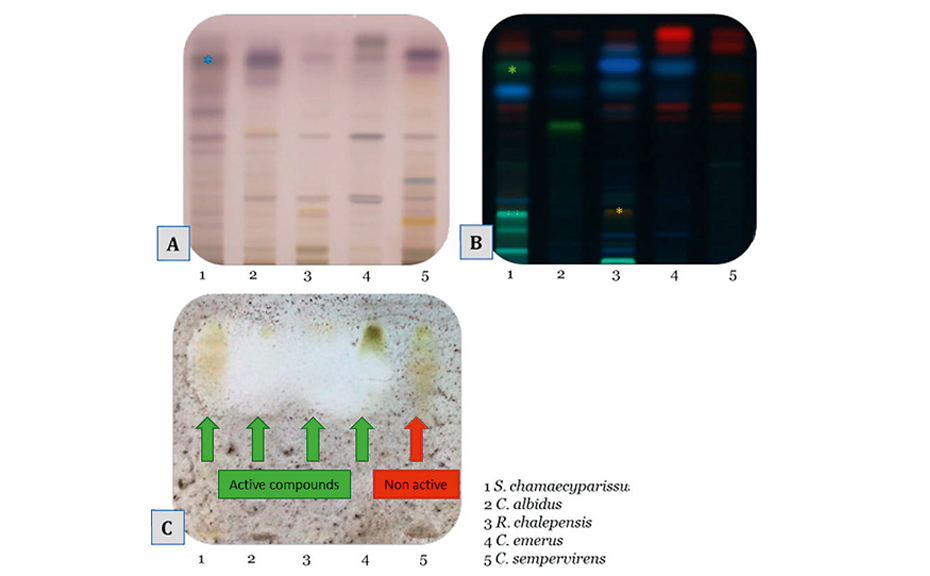
The antifungal capacity of five Mediterranean plants (Santolina chamaecyparissus L., Asteraceae, flowering top; Cistus albidus L., Cistaceae, branches; Ruta chalepensis L., Rutaceae, top; Coronilla emerus L., Fabaceae, flowering top; Cupressus sempervirens L. Cupressaceae, branches) was studied using A. brasiliensis to discover new natural preservatives for the cosmetic industry. First, the raw extracts were screened following the dilution method in 96 well plates.
Afterwards, in order to determine the family of compounds responsible for the antifungal activity, HPTLC bioautography was performed on the extracts.
Except for C. sempervirens, the tested plants showed antifungal activity on the plate. Positive zones are located on top of the plates where unpolar compounds migrate. Terpenoids (purple zones after derivatization with anisaldehyde) and unpolar polyphenols (greenish-yellow zones after derivatization with NP-PEG) seem to bear the antifungal activity. C. sempervirens L. has no antifungal property as shown on the dilution assay.
Further investigations [2] on Santolina chamaecyparissus L. extract led to the isolation and NMR identification of the active compound spiroketalenol ((5S,7Z)-7-(hexa-2,4-diyn-1-ylidene)-1,6-dioxaspiro [4.4]nona-2,8-dien-4-yl acetate). Therefore, Santolina extract seems to be a good candidate as a new natural preservative for cosmetics.
[1] G. Morlock et al. J Chromatogr A 1217 (2010) 6600
[2] A. Kerdudo et al. C R Chimie 19 (2016) 1077
Further information is available online and on request from the authors.
Contact: Prof. Xavier Fernandez, MVBV, Institut de Chimie de Nice, 28 avenue Valrose, 06108 Nice Cedex 2, France, xavier.fernandez@univ-cotedazur.fr