Dr. Wilmer H. Perera serves as Lab Manager at CAMAG Scientific, Inc. in Wilmington, NC. He has collaborated closely with Dr. Jane Manfron’s group in the development of HPTLC methods for the quality control of Brazilian herbal drugs. Herein, he transferred the regular HPTLC method for discriminating Monteverdia ilicifolia from adulterants into an HPTLC PRO method to improve the efficiency of the process. Dr. Jane Manfron holds the position of Associate Professor at the State University of Ponta Grossa, Brazil, and leads the Pharmacognosy Laboratory. Her research focuses on the identification of plant species, using morphology and microscopy. In addition, she studies the chemistry and biological effects of essential oils. Her team involved in this research are Kevin A. Antunes, MSc., Luciane M. Monteiro, MSc., Vera L.P. dos Santos, Ph.D., and collaborators Gustavo Heiden, Ph.D. and Ernestino de S.G.G, Ph.D. from Embrapa Clima Temperado, Pelotas, Rio Grande do Sul, Brazil.
Introduction
The species Monteverdia ilicifolia (Mart. ex Reissek) Biral, also known as espinheira santa, is one of the most commercialized species in Paraná State, Brazil. It is widely used to treat gastritis and gastric ulcers. Despite its popularity, the market is plagued by issues of low-quality products. Common adulterants include Citronella gongonha (Mart.) R.A. Howard, Jodina rhombifolia (Hook. & Arn.) Reissek, Sorocea bonplandii (Baill.) W.C. Burger et al., Zollernia ilicifolia (Brongn.) and Monteverdia aquifolia [1]. Studies have shown that several herbal samples sold in the market have been misidentified and M. aquifolia is frequently sold as M. ilicifolia [1]. An HPTLC PRO method based on the fingerprint of flavonoids and phenolic acids was developed for the discrimination of all relevant herbal drugs and applied for the analysis of commercial samples.
HPTLC is considered the gold standard for the identification of botanicals in many countries. The fully automated version of the technique, HPTLC PRO, brings more hands off during the analysis. The capabiliy to regulate the gas phase during chromatography, using the HPTLC PRO Module DEVELOPMENT, improves the separation efficiency when compared to conventional HPTLC performed by development in the ADC 2.
Standard solutions
Epicatechin was prepared at 28 μg/mL, quercetin and chlorogenic acid at 200 μg/mL, and rutin at 400 μg/mL in methanol. The Universal HPTLC mixture (UHM; prepared in house) was used as a system suitability test (SST).
Sample preparation
5.0 g of leaves of Monteverdia ilicifolia, adulterants and herbal products were milled and extracted with 50.0 mL of methanol by sonication for 15 min at room temperature. The solution was filtered over cotton and then dried using a rotatory evaporator to afford 10 mg of the extract. The evaporated extract was dissolved in a suitable amount of methanol to yield 10 mg/mL.
Chromatogram layer
HPTLC plates silica gel 60 F254 (Merck), 20 x 10 cm are used.
Sample application
5.0 μL of sample and standard solutions, and 2.0 μL of the UHM are applied as bands with the HPTLC PRO Module APPLICATION, 15 tracks, band length 8.0 mm, distance from the left edge 20.0 mm, track distance 11.4 mm, distance from the lower edge 8.0 mm. The first rinsing step (bottle 1 solvent) is done with methanol – acetonitrile – isopropanol – water – formic acid 250:250:250:250:1 (V/V) and the second rinsing step (bottle 2 solvent) is done with methanol – water 7:3 (V/V).
Chromatography
The HPTLC PRO Module DEVELOPMENT used the following parameters: pre-drying for 30 s, activation at 33% relative humidity for 10 min using a saturated solution of magnesium chloride, conditioning with n-butyl acetate – methanol – water – formic acid 15:4:2:2 (V/V) at a pump power of 25% from 50 to 70 mm developing distance, development with n-butyl acetate – methanol – water – formic acid 15:4:2:2 (V/V) to the migration distance of 70 mm (from the lower edge), followed by drying for 5 min [2].
Post-chromatographic derivatization
Plates are derivatized using the HPTLC PRO Module DERIVATIZATION by heating the plate at 100 °C for 180 s then spraying 1.5 mL of Natural Product reagent (NP) (1,0 g of 2-aminoethyl diphenylborinate in 100 mL of methanol) under reduced pressure with nozzle 1 at spraying level 3. A second derivatization was performed by using 1.8 mL of anisaldehyde reagent (add slowly 10.0 mL of acetic acid and 5.0 mL of sulfuric acid to 85.0 mL of icecooled methanol, mix, cool to room temperature, add 0.5 mL of p-anisaldehyde) under reduced pressure with nozzle 2 at spraying level 2, then the plate is heated at 100 °C for 90 s.
Documentation
Images of the plate are captured with the TLC Visualizer 2 in UV 254 nm after development, UV 366 nm after derivatization with NP reagent, and white light after subsequent derivatization with anisaldehyde reagent.
Results and discussion
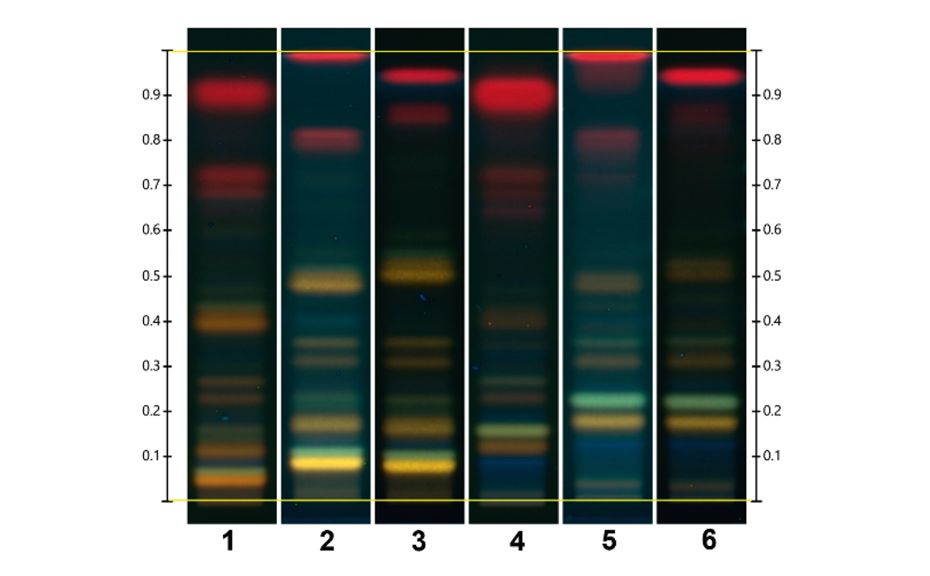
The fingerprint obtained from Monteverdia ilicifolia and related species in the ADC 2 is compared to the fingerprint obtained from the HPTLC PRO System as shown in the figure below. The RF position of all zones obtained with the HPTLC PRO System appears to be higher than with the ADC 2. When no preconditioning or conditioning is used, the chlorophylls migrate with the solvent front but with conditioning all zones are observed between the application position and the solvent front.
Using the HPTLC PRO System a slight separation improvement is observed in the lower part of the chromatogram, below RF 0.4.
Using the HPTLC PRO System with conditioning, the fingerprint of M. ilicifolia is different from those of the potential adulterants including the related species, considering both detection modes. UV 366 nm after derivatization with NP reagent is the most suitable detection mode for identification. In the SST, the UHM generates three main quenching zones in shortwave UV(254 nm) at RF 0.15±0.01, 0.47±0.01, and 0.82±0.01.
Nine samples purchased on the Brazilian market and one online in the USA were analyzed using the developed method. The fingerprints of the samples on tracks 8, 12, 13, 15 and 16, matched that of the M. aquifolia BRM on track 5. The sample on track 9 also displayed a fingerprint of M. aquifolia, but with a couple of additional blueish zones at RF 0.26 and 0.42, indicating the presence of adulterants. The sample on track 11 exhibited a distinct fingerprint, including these blueish zones. This fingerprint had been previously identified as Sorocea bonplandii [1] implying that the sample on track 9 with a similar M. aquifolia fingerprint has also been adulterated with S. bonplandii. The sample on track 10 revealed a clear fingerprint similar to that of the M. ilicifolia BRM while the sample on track 14 also shows similarities to M. ilicifolia with a fainter zone of the flavonoid polyglycosides at RF~0.09. The sample on track 17 exhibited a few zones characteristic of Jodina rhombifolia, although other zones not associated with any adulterants are also observed. Only two of the ten samples match the fingerprint of M. ilicifolia. Although M. ilicifolia and M. aquifolia thrive in different habitats, the misuse of M. aquifolia for M. ilicifolia in commercial samples is very common. HPTLC PRO proves to be an effective approach to rapidly discriminate M. ilicifolia from its adulterants in commercialized samples using the flavonoid fingerprint.
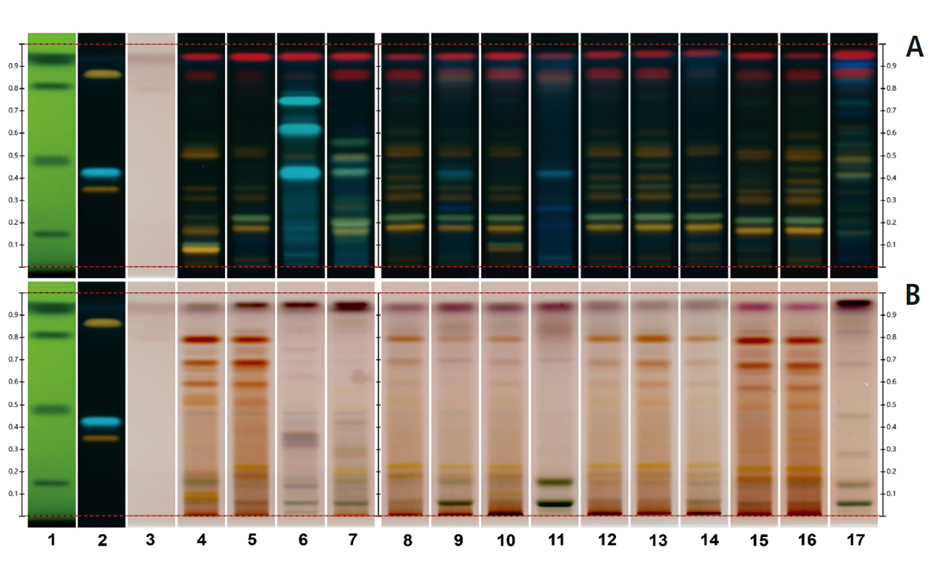
Chromatograms of the UHM in UV 254 nm (track 1), rutin, chlorogenic acid, and quercetin in UV 366 nm after derivatization with NP reagent (track 2, with increasing RF ), epicatechin 28 μg/mL post derivatization with anisaldehyde on top of NP reagent (track 3), Monteverdia ilicifolia botanical reference material (BRM) (track 4), Monteverdia aquifolia BRM (track 5), Citronella gorgonha BRM (track 6), Jodina rhombifolia BRM (track 7) and herbal products tested from Brazilian market (tracks 8–16) and from USA (track 17).
[1] Antunes KA et al. Microsc Microanal (2023) https://doi.org/10.1093/micmic/ozad098
[2] Antunes KA et al. Nat Prod Res, accepted (2023)
Further information is available on request from the authors.
Contact: Dr. Jane Manfron, Postgraduate Program in Pharmaceutical Sciences, State University of Ponta Grossa, 4748 Carlos Cavalcanti Avenue, 84030-900 Ponta Grossa, PR, Brazil. janemanfron@hotmail.com
Dr. Wilmer H. Perera, CAMAG Scientific, Inc., 515 Cornelius Harnett Drive Wilmington, NC 28401, USA, wilmer.perera@camag.com